Clik here to view.

Yao et al. found that certain brain enhancers were functionally conserved between mice (left) and acorn worm (right), despite very limited sequence conservation.
Douglas Epstein
A study published this week in Nature Genetics shows that enhancers can be conserved across very long evolutionary distances, even without extensive sequence conservation.
Understanding how non-coding elements in the genome, such as enhancers, are recognized by regulatory proteins is key to understanding how cells and tissues coordinate their specific gene expression programs. One way to approach the problem is to identify homologous elements that are evolutionarily conserved among distant relatives. This has generally been done by searching for non-coding regions with conserved DNA sequences, which suggests that the elements are functional. However, recent research has found that sequence conservation doesn’t always explain functional conservation of regulatory elements. For example, Michael Eisen and colleagues found that enhancers of the developmental gene even-skipped are functionally conserved between scavenger flies (in the genus Sepsidae) and Drosophila, even though they weren’t highly conserved at the sequence level. Deep functional conservation of enhancer elements in Drosophila was also experimentally shown by Alexander Stark and colleagues.
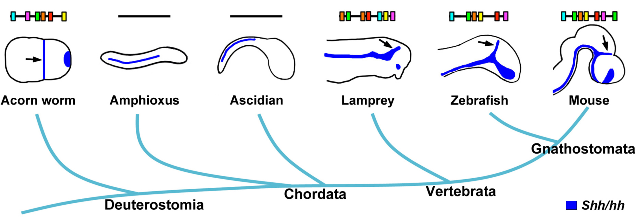
These and other studies demonstrated that overall sequence conservation is not necessary for functional conservation of regulatory elements, but how deep can this conservation go? Drosophila and scavenger flies share very similar body plans and developmental programs. But can organisms with divergent body plans still share functionally conserved elements in the absence of overt sequence conservation? A new paper by Douglas Epstein and colleagues finds that enhancers that drive expression of the Sonic hedgehog gene (Shh) in the zona limitans intrathalamica (zli) region of the brain are more ancient than chordates. The enhancers are functionally conserved in animals as distantly related as mice and the hemichordate Saccoglossus kowalevskii, even though they are not conserved at the DNA sequence level and even though the hemichordate does not have a recognizable zli.
The authors searched for enhancers regulating similar spatiotemporal patterns of expression to a known Shh brain enhancer, SBE1, active in the zli. They used a computational approach to identify six short motifs that were significantly enriched in this set of enhancers. The motifs, ranging in size from 6–14bp, were not found in any consistent order within the SBE1-like enhancers, but as long as they were present in some order, their function was conserved within vertebrates.
Clik here to view.
Yao et al. 2016 Nature Genetics
They then looked for more ancient conservation of the elements. For this, they used the hemichordate Saccoglossus kowalevskii (also known as the acorn worm), which shows similar patterns of gene expression during development to vertebrates, but that doesn’t have a distinct zli. However, it does express hedgehog (hh) in a narrow band of cells in the embryo that, based on the surrounding patterns of gene expression, appears to be zli-like. They found a region in an intron of hh that contained the six motifs they found in vertebrates, without any other sequence homology. Using transgenic experiments in mouse and S. kowalevskii embryos, they showed that a mouse SBE1 enhancer could drive hh expression in the zli-like region in the acorn worm embryos, and the acorn worm SBE1-like element could drive expression in the mouse zli.
We asked the corresponding author of the study, Douglas Epstein, to tell us a little more about the study:
Why did you choose to focus your study on enhancers active in the zona limitans intrathalamica (zli)?
This study was motivated by our longstanding interest in determining how genes are regulated during brain development. The zli fit the bill due to its notable role as a source of the secreted morphogen protein Sonic hedgehog (Shh) that patterns the thalamic and prethalamic territories. Also, SBE1 was the first Shh enhancer that I had identified during my postdoc, but I set it aside for many years to work on other projects. It was only after Eddy Rubin introduced me to the VISTA Enhancer Browser during a visit to Penn that our curiosity was piqued. This database of experimentally validated enhancers contained numerous examples of reporter constructs active in the zli, with similar specificity to SBE1. This intriguing observation renewed our interest in the zli and was the impetus behind our current publication in Nature Genetics.
You used both mice and the hemichordate S. kowalevskii for your studies. What is the significance of the data from the hemichordate studies, and how does it further our understanding of vertebrate biology?
Our comparative approach was useful for revealing the evolutionary history of SBE1 and demonstrating that the regulatory mechanisms used to build components of the vertebrate brain show evidence of deep homology, a term used by Shubin, Tabin and Carroll in their 2009 Nature review article to describe the “ancient similarity in patterning mechanisms in diverse organisms”. It’s fascinating to think that at least some of the design principles for building the complex vertebrate brain were taken from a shared ancestor with the humble acorn worm.
Did you expect to find so little conservation at the DNA sequence level? Were you surprised to find that the enhancer elements were still functionally conserved? Why or why not?
On all levels there was quite a bit of surprise that the regulatory logic is conserved and functional in such dramatically different manifestations of the ectoderm in mice and hemichordates. Once we found that the set of binding sites in SBE1-like enhancers did not obey any specific rules in terms of order or spacing, we realized that SBE1 homologues or orthologues in other species may not share much, if any, sequence conservation. There was precedent for this in the literature from studies in Drosophila and zebrafish. When Chris Lowe (a co-author of the study) and I first started to collaborate in 2012, he was particularly jaded (his words) by the attempts of other groups to discover conserved enhancers in hemichordates by computational means. He felt that the state of the field is not to expect this level of regulatory conservation between phyla and especially between phyla with such divergent body plans. Chris also claims that I was unfettered by this burden (his words) and upon finding the transcription factor binding sites, was convinced that the experiments were worth pursuing. This last point can’t be overstated. Had we not first determined the cis-regulatory signature of SBE1-like enhancers we would not have found mmSBE5 (a mouse homologue of SBE1) or skSBE1 (the SBE1 element from S. kowalevskii).’
Your study included an experiment in which you test the activity of mammalian SBE1 and SBE5 in the hemichordate system. I assume this must have been very technically challenging. How did you overcome these challenges? Were the results what you expected?
Yes, it was challenging to develop the transgenics in hemichordates. Co-author Paul Minor largely followed the Krumlauf lab’s efforts on lamprey and one of the biggest hurdles was finding the right endogenous promoter that would work. He tried several and gbx was the one that was the most effective. Another obstacle in working with Saccoglossus is the restricted breeding season. Most of the embryo injections could only be done in September during the peak spawning season.
How do you see your findings impacting on future research?
In terms of our research, the interesting part to figure out next is the function of the Hedgehog (Hh) protein in the zli-like ring at the boundary of the proboscis-collar region of hemichordate embryos. We’d love to use CRISPR-Cas9 to delete skSBE1 and determine whether the loss of Hh disrupts some aspect of neuronal patterning or connectivity. Mining the S. kowalevskii genome for other examples of deep homology in regulatory circuits involved in brain development should also be informative. We also plan to determine whether the small deletion of SBE5 causes behavioral abnormalities in mice due to thalamic dysfunction. In terms of the broader impact of our work, I hope that it inspires even more rigorous experimental approaches to the analysis of enhancer function and evolution.
Click here for access to a free-to-read PDF of the paper